The end
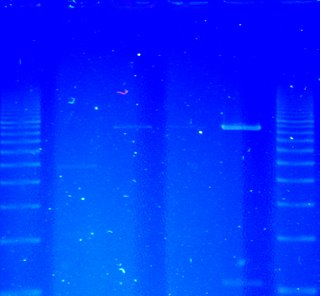
Now I am done. I leave in a few hours for Botswana and South Africa - perfect timing. If I could find a source of isocitrate, I could do a functionality and specificity assay, but frankly I am satisfied with the protein on the gel. Yay! And I budgeted fairly well, using up most everything but some media and yeast genomic DNA preps. I did go over budget, spending $1100. Oh well. It was fun.
Thanks again to Mr. Rene Gillibert, Ms. Marietta Dunaway, Mr. Matthew Hamilton, Dr. Fang, Ms. Vicky Kim of Novagen, Ms. Janet Long of John Muir Regional Medical Center, Mr. Dunaway, the tech people at Epicentre, the sales people at Epicentre for offering a discount, and to the sales people at USB for offering a discount.
Thanks again to Mr. Rene Gillibert, Ms. Marietta Dunaway, Mr. Matthew Hamilton, Dr. Fang, Ms. Vicky Kim of Novagen, Ms. Janet Long of John Muir Regional Medical Center, Mr. Dunaway, the tech people at Epicentre, the sales people at Epicentre for offering a discount, and to the sales people at USB for offering a discount.